Omics Workflows
Omics Workflows enables processing genomics data using user defined workflows described in WDL or Nextflow. A single invocation of a workflow is called a run, and a single process within a run is called a task. Users have to upload a zip file containing the Workflow definition file(s) and Omics Workflows will automatically provision and scale the underlying infrastructure for running their bioinformatics workflow. A workflow can be run on genomics data stored in an Amorphic dataset or in an Omics Sequence store.
To use Omics Workflows, HCLS should be enabled in the environment.
How to create an Omics Workflow?
- Click on
+ New Omics Workflow - Upload a zip file containing the Workflow definition file(s).
- Fill in the required fields (Details listed below)
| Properties | Details |
|---|---|
| Workflow Name | A name for the Omics Workflow. Workflow Name should only contain 1-128 alphanumeric, - & _ characters. |
| Workflow Engine | The desired workflow definition engine: WDL or Nextflow |
| Description | Description of the workflow being created. |
| Main | An optional attribute which specifies which file should be the entry point for workflow execution. |
| Storage Capacity | Desired storage capacity for the workflow, can be between 1-20,000 GB. |
| Keywords | Keywords indexed & searchable in app. Choose meaningful keywords to flag related workflows & easily find them later. |
| Dataset Read/Write Access | Specifies which all datasets the workflow needs Read/Write access to respectively. |
Atmost 1 dataset can be specified for write access. If it is specified, worklow output will be written to this dataset.
Below image shows how to create a new Omics Workflow:

Once a workflow is created ,the details page will be as shown below:

How to run an Omics Workflow
- Select the Omics Workflow that you want to run and go to
RUNStab. - Click on
+ Trigger Run - Fill in the required fields (Details listed below)
| Properties | Details |
|---|---|
| Run Name | A name for the Omics Workflow Run. Workflow Run Name should only contain 1-128 alphanumeric, - & _ characters. |
| Storage Capacity | Desired storage capacity for the workflow run, can be between 1-20,000 GB. |
| Log Level | Required log level for the workflow run. Can be any of OFF, FATAL, ERROR or ALL. |
| Run Parameters | The parameters required to run the workflow. |
Below image show how to start Omics Workflow Run:

Below image show Omics Workflow Runs:
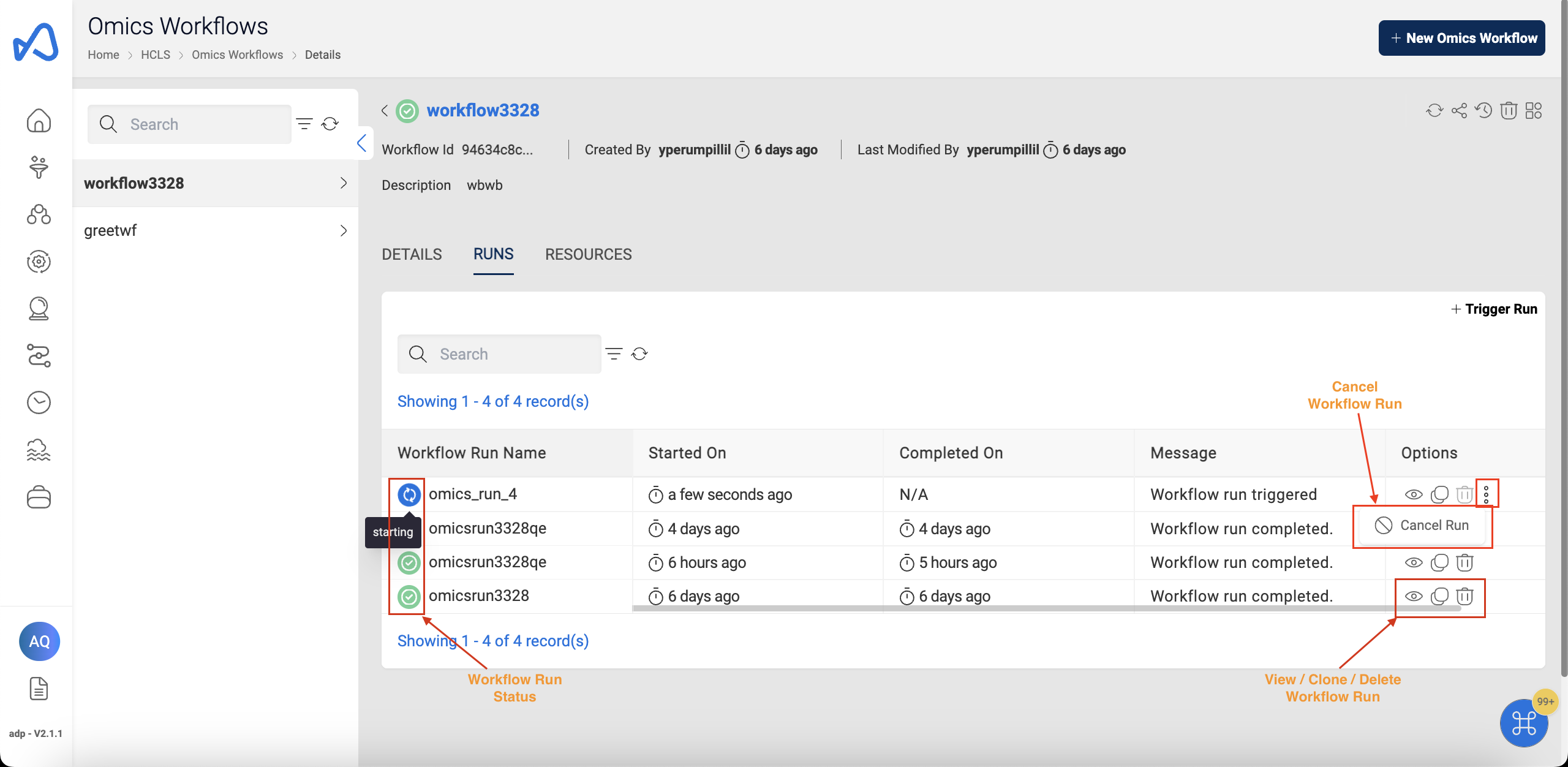
You can cancel a Workflow Run while it is in Pending, Starting or Running state.
How to download Omics Workflow Run logs
- Click on the Omics Workflow Run you want the logs for.
- Click on
Generate Logsbutton. - After the successful generation of logs, you can download logs of respective runs by clicking
Download Logsbutton.
Below image show how to download Omics Workflow Run Logs:

You can view details of each tasks of an Omics Workflow Run and download logs of each tasks by going to the Run Tasks section of an Omics Workflow Run.